Tuesday A: Getting to know the variational application¶
In today’s tutorial we will build on the first steps taken yesterday and learn more about how to run the FV3-JEDI variational data assimilation application and how to modify it. First we will learn how to assess whether a particular run is healthy and then investigate some ways to change the run to improve the analysis.
In summary in this activity you will:
Learn how to check for healthy data assimilation runs in JEDI.
Get to know the variational data assimilation application.
Learn how to alter the behavior of the variation assimilation by adjusting the number of iterations.
Step 1: Download the data¶
In order to build on the work done yesterday we need some different data. This data will be the basis for this tutorial and the other tutorials covering FV3-JEDI over the week. Begin by downloading and extracting this data to your tutorials directory:
cd ~/jedi/tutorials
wget https://fv3-jedi-public.s3.amazonaws.com/AcademyNov2020/20201001_0000z.tar
tar -xvf 20201001_0000z.tar
This directory contains everything needed to run a data assimilation application with JEDI. The
files in this directory are designed to cover the 6 hour assimilation window that goes from
2020-09-30T21:00:00 to 2020-10-01T03:00:00. The name of the tar file and directory denotes the
central time of the window. Take a minute to look around the 20201001_0000z/Data directory
and familiarize yourself with the kinds of files that are required in order to run the data
assimilation application. You’ll see background files through the window, ensemble files through the
window for a 32 member ensemble, observations, CRTM coefficient files and some configuration files
needed by FV3-JEDI. Today we will focus on the 3DEnVar assimilation run and so will use data valid
at the center of the window, 2020-10-01T00:00:00
Step 2: Revive your environment¶
Before anything can be run we need to revive the parts of the environment that are not preserved when the instance in stopped and restarted.
Begin by entering the container again:
cd ~/
singularity shell -e jedi-gnu-openmpi-dev_latest.sif
Your prompt should now look something like:
Singularity>
Once in the container be sure also to remove limits the stack memory to prevent spurious failures:
ulimit -s unlimited
ulimit -v unlimited
If you didn’t already do this you need to install the fv3-jedi-tools package to provide the diagnostics:
cd ~/jedi
git clone https://github.com/jcsda-internal/fv3-jedi-tools
cd fv3-jedi-tools
/usr/local/miniconda3/bin/pip install --user -e .
Even if FV3-JEDI-TOOLS was installed previously the path still needs to be set in each session:
export PATH=$HOME/.local/bin:$PATH
Step 3: Run a 3DEnVar analysis for the C24 (4 degree model)¶
In the previous days tutorial you ran using a script that produced the entire run. Today we will manually run each step in order to see all the steps involved and more easily re-run specific steps after modifying the yaml file.
For simplicity make a convenience pointer to the place where JEDI is built:
JEDIBUILD=~/jedi/build/
Data assimilation involves a background error covariance model, which we will learn more about in the upcoming lectures on SABER and BUMP. The variational assimilation system we are running today uses a pure ensemble error covariance model but it requires localization in order to give a sensible analysis. The first step we need to take is to generate this localization model. Do not worry about understanding the configuration in this step, we will learn more about it later this week. To generate the localization model enter the following:
# Move to the directory where the executable must be run from
cd ~/jedi/tutorials/20201001_0000z
# Create a directory to hold the localization model files
mkdir -p Data/bump
# Run the application to generate the localization model
mpirun -np 6 $JEDIBUILD/bin/fv3jedi_parameters.x Config/localization_parameters_fixed.yaml
The localization model is fixed and does not even depend on the time for which the analysis is being run. So the above step is taken only once, even if changing the configuration for the variational assimilation.
Now we can run the data assimilation system and produce the analysis.
# Create directory to hold the resulting analysis
mkdir -p Data/analysis
# Create directory to hold resulting hofx data
mkdir -p Data/hofx
# Create directory to hold resulting log files
mkdir -p Logs
# Run the variational data assimilation application
mpirun -np 6 $JEDIBUILD/bin/fv3jedi_var.x Config/3denvar.yaml Logs/3denvar.log
Step 4: Check that we produced a healthy looking analysis¶
There are a number of diagnostics that can be useful for determining whether an assimilation run was healthy. That is to say that is converged to within some criteria and produced a sensible analysis and fit to observations. The diagnostics we’ll explore in this tutorial are:
Plotting hofx [\(h({x})\)] and observation difference maps.
Plotting the convergence statistics.
Plotting the observation fit ‘innovation’ statistics.
Plotting the increment.
In the first practical we plotted the increment and hofx maps through the script. Configuration yaml
files are provided to make similar plots for this experiment in the /Diagnostics directory
and the plots will be generated manually.
First off we can generate the maps for the \(h({x})\) data using:
fv3jeditools.x 2020-10-01T00:00:00 Diagnostics/hofx_map.yaml
This will produce the hofx map plot for air temperature from the aircraft observations.
Warning
This and other diagnostics will create a figure file. This file is located in the same directory
as the file read to produce the diagnostic. These paths can be found in the configuration files
used to create the diagnostic e.g. Diagnostics/hofx_map.yaml.
You can try uncommenting and adjusting the values of colorbar minimum and
colorbar maximum in the yaml file to produce a plot with better scaling. Now try adjusting
the yaml to have variable: air_temperature@ombg and plotting again. You’ll need to choose
the colorbar limit options in the configuration. This plot will show you the difference
between the observation values and the background simulated observations [\(h({x_b})\)]. Now try
plotting variable: air_temperature@oman, this will give you the difference between the
observation values and the analysis simulated observations [\(h({x_a})\)]. You should notice
some of the larger values disappear from the oman plot compared to the ombg plot. This is as
expected, the analysis has moved the state closer to the observations.
That the assimilation draws closer to the observations can also be verified quantitiaively by
examining the log file produced by the run. This file is located at Logs/3denvar.log.
First of all you can search in this file for the string Jo/n. This will reveal lines like:
CostJo: Nonlinear Jo(Aircraft) = 238308, nobs = 268090, Jo/n = 0.88891, err = 2.19395
CostJo: Nonlinear Jo(AMSUA-NOAA19) = 53002.4, nobs = 60049, Jo/n = 0.882653, err = 2.00229
In this example the value Jo/n = 0.88891 shows the fit to aircraft observations before the
analysis and Jo/n = 0.882653 the fit to the AMSU-A observations before the analysis. By continuing
to search through the file you will find values for Jo/n after the analysis. Here you’ll see
lower values for each instrument, indicating a closer fit to the observations. Jo refers to the part
of the cost function that measures the difference between the state and the observations. It is
normalized by n, the number of observations.
The log file contains lots of other information about the convergence, but it is best to visualize this information to check the convergence, rather than looking through the log file. fv3-jedi-tools has the ability to create these diagnostics using the following:
fv3jeditools.x 2020-10-01T00:00:00 Diagnostics/da_convergence.yaml
This will produce five diagnostic plots showing aspects of the cost function. The key plot to look
at is the one labelled drpcg-normalized-gradient-reduction. In a healthy data assimilation
run the normalized gradient reduction will decline and then flatten off with some relatively small
value. The plot produced should look similar to this:
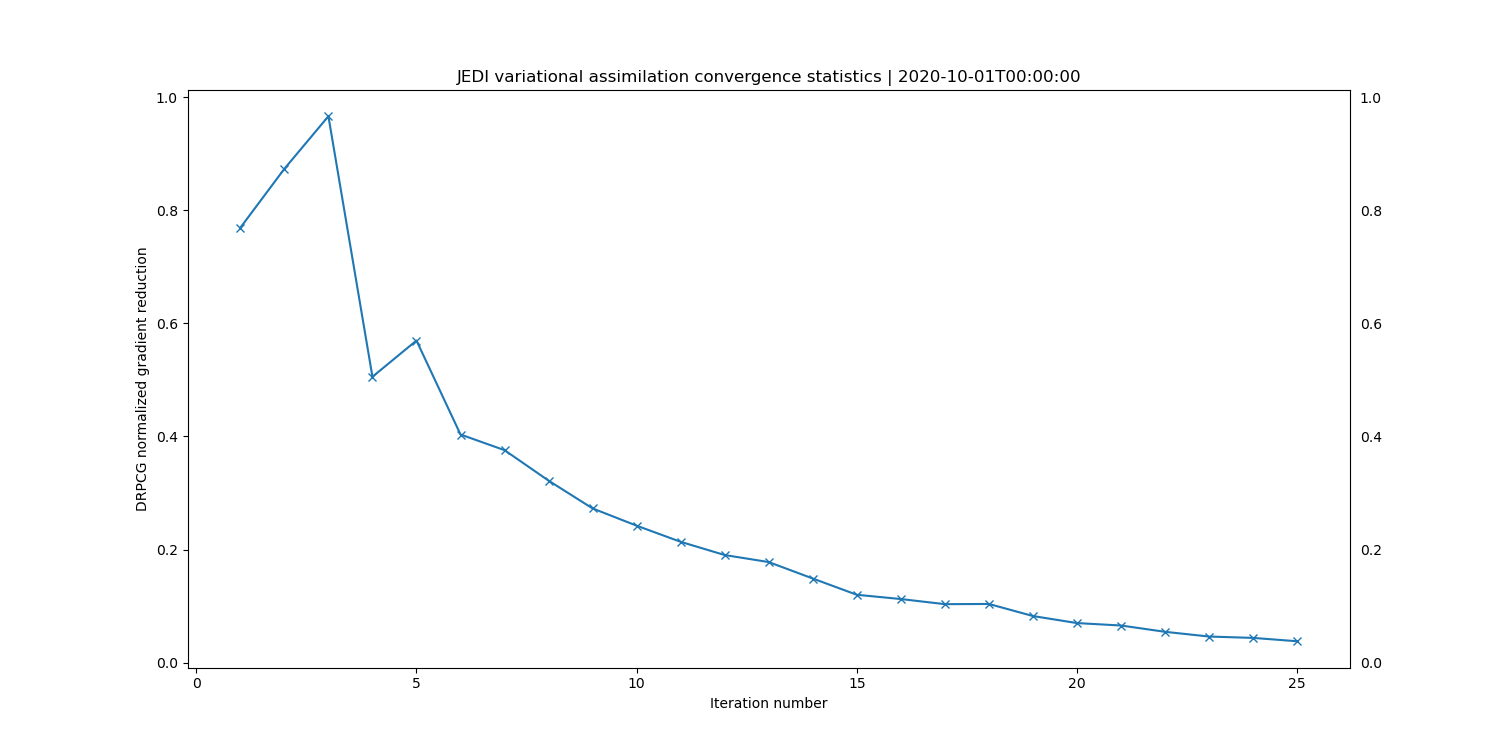
Initially the gradient reduction increases, but after 25 iterations or so the change from one iteration to the next is quite small, showing that the cost function is approaching its minimum, but perhaps not completely converged yet. Note that this metric is normalized with respect the the value at the beginning of the outer loop, so values are close to 1 at the beginning and then should reduce. In a typical operational weather forecast system values of the order of \(10^{-2}\) would be sufficient to indicate convergence. Though in most cases a maximum number of iterations would be satisfied before the value of the reduction.
As mentioned above the assimilation should result in an analysis state closer to the observations than the background state was. While this is most readily verified by looking at the log, it is often useful to view this in a statistical sense as well. This can be done with the following:
fv3jeditools.x 2020-10-01T00:00:00 Diagnostics/hofx_innovations.yaml
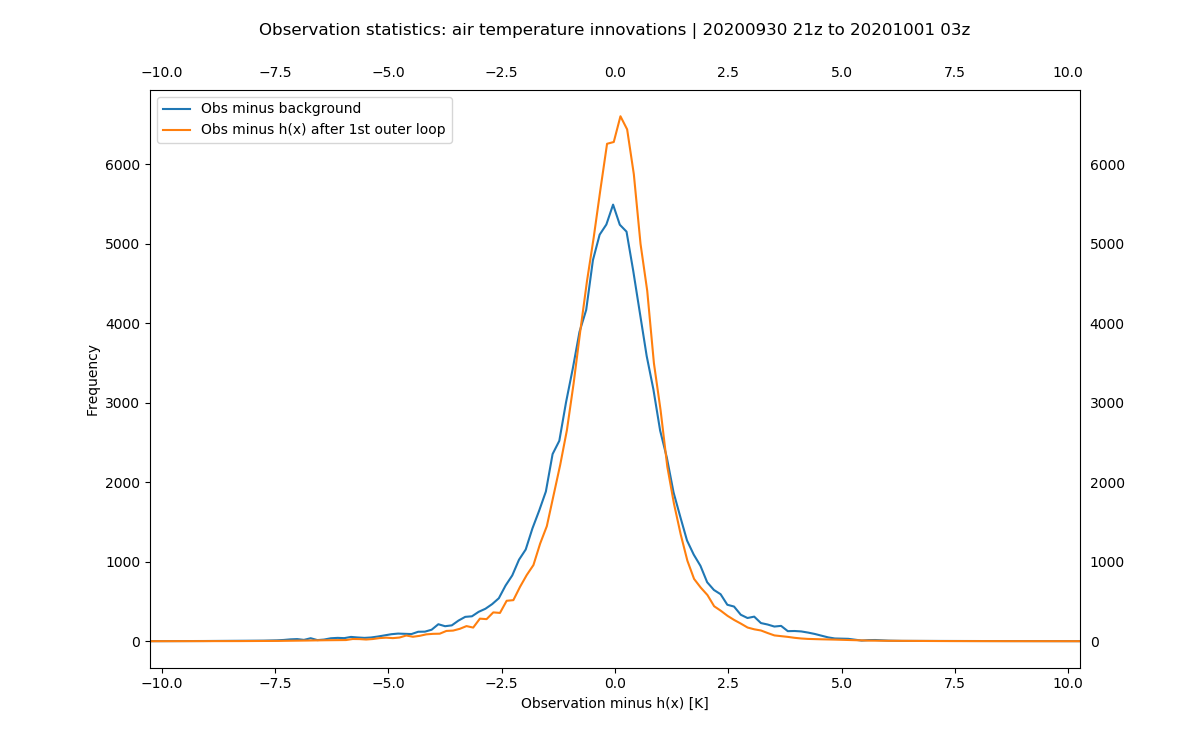
The resulting plot (in Data/hofx/) should look similar to the below plot, with two probability distributions. In
this figure the blue curve shows the probability for the observation minus observation simulated
from the background and the orange curve the probability for the observation minus observation
simulated from the analysis. The distribution has less spread and a mean closer to zero for the
analysis. In Diagnostics/hofx_innovations.yaml there’s an option called
number of bins, which determines the number of bins to use when creating the histograms,
which are in turn interpolated to give the probability distributions. Try adjusting this number to
see how the figure can change. If you decide to look at the AMSUA data you may need to use fewer
bins to avoid noisy looking plots.
As in the first day practical you can plot the increment produced by the assimilation. The first
step is to generate the increment using the diffstates application as follows:
mkdir Data/increment
mpirun -np 6 $JEDIBUILD/bin/fv3jedi_diffstates.x Config/create_increment_3denvar.yaml
Once it has been created it can be plotted with:
fv3jeditools.x 2020-10-01T00:00:00 Diagnostics/field_plot.yaml
You can try adjusting the variable and level to be plotted in Diagnostics/field_plot.yaml.
Note that you can do ncdump -h Data/increment/inc.3denvar.20201001_000000z.nc4 to see
which fields are included in the analysis.
Step 5: Improve the analysis¶
Before continuing, make a backup of the data assimilation configuration file. In later tutorials we will use it as the starting point for other experiments.
cp Config/3denvar.yaml Config/3denvar_backup.yaml
If you open Config/3denvar.yaml you will see all the set up for the assimilation run. Scrolling to the bottom of this file you will find a section that looks like:
variational:
# Minimizer to employ
minimizer:
algorithm: DRIPCG
iterations:
- ninner: 25
gradient norm reduction: 1e-10
test: on
# Increment geometry
geometry:
trc_file: *trc
akbk: *akbk
layout: *layout
npx: *npx
npy: *npy
npz: *npz
fieldsets:
- fieldset: Data/fieldsets/dynamics.yaml
- fieldset: Data/fieldsets/ufo.yaml
# Diagnistics to output
diagnostics:
departures: ombg
This part of the yaml controls the minimization of the cost function in the assimilation system.
Variational data assimilation can consist of outer loops and inner loops. In the case show here
one outer loop is used and there are 25 inner loops, denoted by the line ninner: 25. Inner
loops essentially refer to the number of iterations taken to minimize the cost function. Between
each outer loop the increment is added to the background and the cost function is re-linearized. As
such a single outer loop with say 50 inner loops will produce a different result to using two outer
loops with 25 inner loops each.
As we saw in some of the diagnostics above the assimilation system was not that well converged. Try
adjusting to ninner: 100 in the yaml file. Then go back and run the assimilation again, the
final part of Step 3: Run a 3DEnVar analysis for the C24 (4 degree model). With more inner loops now re-examine the diagnostics. How many inner
loops is sufficient to see convergence? Note that running again as before will overwrite any previous
diagnostics so you may wish to move them out of the way for comparison purposes.
Another thing that can be done is to add outer loops to the assimilation. There is no nouter
in the configuration, this is controlled by inserting a list of outer loops in the yaml. Adding
another outer loop in this example would look like:
# Inner loop(s) configuration
variational:
# Minimizer to employ
minimizer:
algorithm: DRIPCG
iterations:
# FIRST OUTER LOOP
- ninner: 25
gradient norm reduction: 1e-10
test: on
geometry:
trc_file: *trc
akbk: *akbk
layout: *layout
npx: *npx
npy: *npy
npz: *npz
fieldsets:
- fieldset: Data/fieldsets/dynamics.yaml
- fieldset: Data/fieldsets/ufo.yaml
diagnostics:
departures: ombg
# SECOND OUTER LOOP
- ninner: 25
gradient norm reduction: 1e-10
test: on
geometry:
trc_file: *trc
akbk: *akbk
layout: *layout
npx: *npx
npy: *npy
npz: *npz
fieldsets:
- fieldset: Data/fieldsets/dynamics.yaml
- fieldset: Data/fieldsets/ufo.yaml
diagnostics:
departures: ombg
Note that with two outer loops you need to adjust to number of outer loops: 2 in
Diagnostics/hofx_innovations.yaml if using that diagnostic. Take a look through all the
diagnostics. Did having a second outer loop help? Try playing with combinations of inner and outer
loops and looking at how the results change.